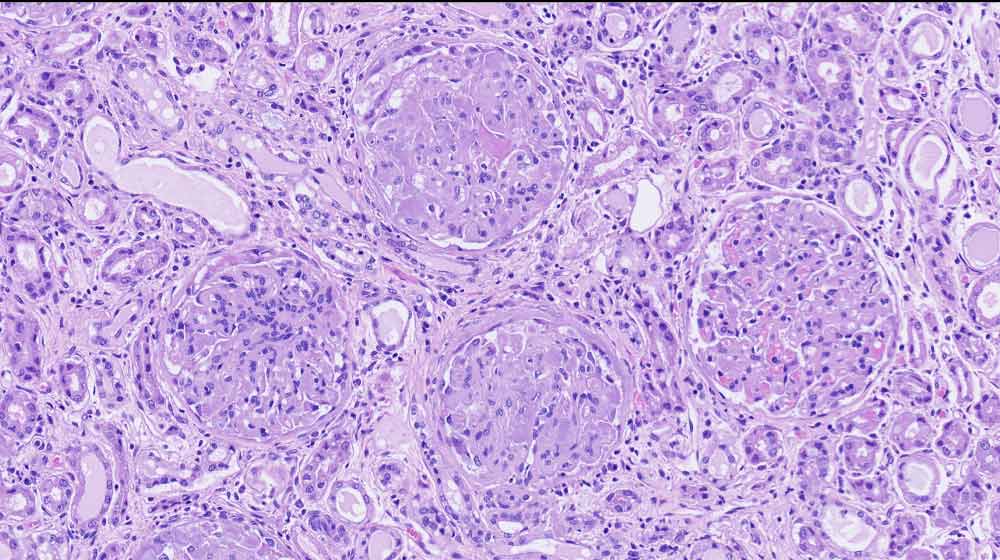
Hereditary transthyretin (TTR) amyloidosis (hATTR) is a rare, life-threatening disorder caused by amyloidogenic coding mutations located in the TTR gene.
To date, more than 130 amyloidogenic mutations have been identified in the coding regions of the TTR gene, which are the cause of the disorder.
Previous data strongly support the role of non-coding regulatory variation on TTR gene expression as one of the mechanisms affecting the phenotypic manifestations observed in carriers of TTR amyloidogenic mutations.
Epigenetic modifications are fundamental mechanisms in modulating a wide range of molecular functions and potential targets to develop novel treatments among genomic regulatory features.
While epigenetic modifications have the potential to be involved in hATTR pathogenic mechanisms, to our knowledge, no study has explored methylation changes of patients affected by this life-threatening disease.
In this recent study, an epigenome-wide association study (EWAS) was conducted assessing more than 700,000 methylation sites and testing the epigenetic difference of TTR coding mutation carriers vs. non-carriers.
Read the original publication of this study here: [Study Identifies Methylation Sites Linked to hATTR]
This study aimed to understand the high phenotypic variability observed among carriers of TTR disease-causing mutations.
Epigenetic profiling of Italian patients identified methylation sites associated with hereditary transthyretin amyloidosis.
Over 700,000 sites in 80 candidates were assessed. A total of 38 symptomatic and 10 asymptomatic patients with TTR mutations were tested and 32 control patients.
The researchers tested the whole blood of the hATTR patients, asymptomatic carriers of TTR mutations, and non-carrier controls.
They compared methylation changes measured as M values, a reference-based method that factored in the cell-type composition’s heterogeneous nature.
Cell proportions for five cell types were detected: B cells, T cells, monocytes, granulocytes, and natural killer cells. A linear regression predictive analysis was also used.
It was found that there were notable methylation changes in a particular gene (BACE2), which is associated with amyloid fiber formation, amyloid-beta metabolism, and Alzheimer’s disease.
Further test results also showed three other significant points:
- TTR and BACE2 both have APP (amyloid-beta precursor protein) as a validated protein interactor.
- Val30Met disrupts a methylation site within the TTR gene region, cg13139646, causing a severe hypomethylation in carriers of this amyloidogenic mutation (standardized regression coefficient 2.18; P = 3.34 × 10–11).
- Cg13139646 showed co-methylation with cg19203115 (Pearson’s r2 = 0.32), which showed significant epigenetic differences between symptomatic and asymptomatic carriers of amyloidogenic mutations (standardized regression coefficient, −0.56; P = 8.6 × 10–4).
The study unearthed new insights associated with the molecular process involved in the intricate heterogeneity of hATTR, underlining the tole of epigenetic regulation in this rare disorder.
Takeaways:
- BACE2 is expressed in tissues affected by the TTR amyloid deposits (i.e., heart, nerves, colon, small intestine, and adipose tissues).
- Accordingly, the methylation change observed in the TTR mutation carriers is possibly due to the role of BACE2 in response to the inflammation induced by the TTR amyloidogenic process in peripheral tissues.
- Due to the small sample size, further studies are needed to distinguish the epigenetic associations’ particular structure.
- “The understanding of how methylation changes modulate the penetrance, and the severity of TTR mutations could lead to the identification of novel targets to develop treatments and screening tools for the carriers,” the authors said.
You can read the original publication of this study here: [Study Identifies Methylation Sites Linked to hATTR]